Home Projects Publications
Proteins play an important role in the living world. Their diversity is huge and fascinating. In our laboratory we study proteins in a complex manner. We are mainly interested in the relationship between the structure and catalytic activity of enzymes, looking for new proteins with interesting properties, optimizing the production of recombinant proteins and developing new molecular modeling methods to simulate slow and computationally demanding processes such as protein folding or interactions of protein with ligands.
Current projets
Cold-active enzymes
Enzymes from organisms living in permanently cold environments (e.g. mountain and polar regions) have significantly higher activity at low temperatures compared to enzymes from meso- and thermophilic sources. This property makes them interesting biocatalysts for low temperature applications. In our group we study cold-active enzymes (specifically chitinase, amylase, cellulase, β-galactosidase), which have a potential for use in the paper, food or pharmaceutical industries.
Enzymes with therapeutical potential
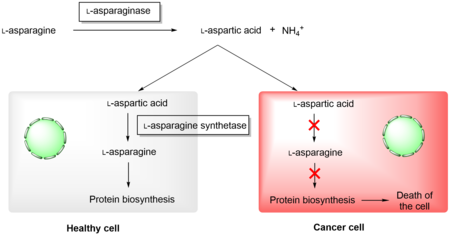
Asparaginases have found application in medicine in the treatment of acute myeloid leukemia. It uses their natural ability to cleave L-asparagine, which is depleted in tumor cells, as a result of which protein biosynthesis is stopped and apoptosis occurs. Healthy cells are able to synthesize L-asparagine due to the presence of L-asparagine synthetase and are therefore not affected by this type of treatment. Preparations containing recombinant L-asparaginase from E. coli and Dickeya dadantii are currently used in practice, however, the application of both enzymes has a number of complications, and therefore it is necessary to look for other sources of L-asparaginase with more suitable properties for clinical applications.

Nucleases have been studied for a long time because of their anti-tumor effects. So far, the main focus has been on animal nuclases. However, in our group we are focusing on enzymes from other sources, such as plants or bacteria. These enzymes are studied using molecular biology and protein engineering methods. Biological studies and crystallographic experiments are carried out in cooperation with the Institute of Plant Molecular Biology, the Institute of Physiology and Animal Genetics and the Biotechnology and Biomedical Center of the Academy of Sciences and Charles University in Vestec (BIOCEV).
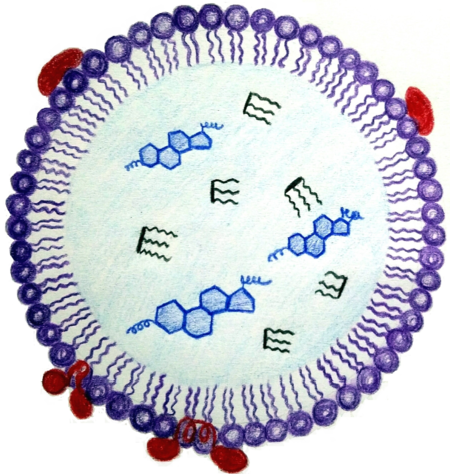
Structural protein of lipid droplets
We are interested in cellular lipid droplets and their life cycle, with special emphasis on the characterization of proteins associated with lipid droplets permanently or within a specific stage of the life cycle. Our goal is to contribute to the description of the life cycle of these specialized organelles and thus to the biotechnological use of both lipid droplets themselves (lipid and biofuel production, oil degradation, drug transport and immobilization systems) and their associated proteins (fusion anchors for industrial recombinant protein expression). As a model organisms we use both prokaryotic (marine bacteria Alcanivorax borkumensis capable to break down oil) and eukaryotic (plant Arabidopsis thaliana and yeast Saccharomyces cerevisiae) organisms.
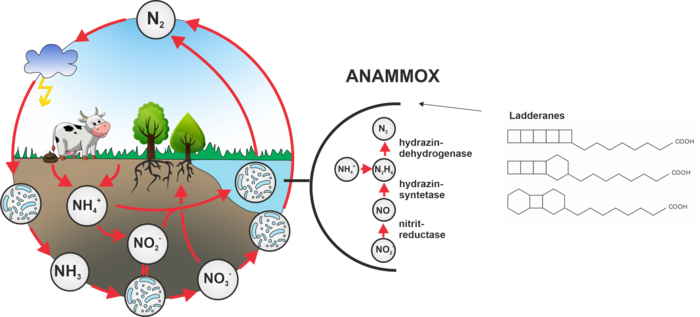
Proteins involved in the synthesis of ladderans in anammox bacteria
So far not well-known anammox bacteria (from the English anaerobic ammonium oxidation) were discovered in the mid-1980s and seem to play an important role in the nitrogen cycle on Earth. Anammox bacteria directly use ammonia as a source of energy, which they convert into molecular nitrogen . They do not need oxygen and, instead of producing carbon dioxide, they consume it. for this purpose they contain an organelle called the anammoxosome, in which energy metabolism is located. The membrane of the anammoxosome is composed of very interesting phospholipids, which have very specific lipids attached – ladderans. During the processing of ammonia in the anammoxosome, a highly reactive and toxic hydrazine is formed as an intermediate, which is just held inside the organelle by ladderans, which, unlike common fatty acids, give the membrane lower permeability to various substances. The biosynthesis of ladderans has not yet been elucidated and is the subject of our research.
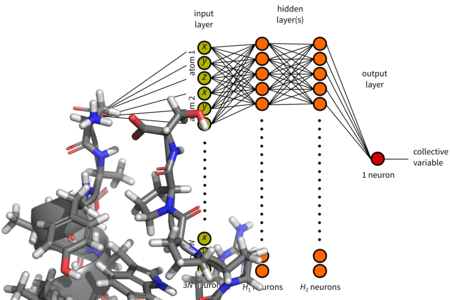
Molecular modelling
Computer simulations make it possible to study complex dynamic behavior of proteins, saccharides, other biomolecules and their assemblies. The main drawback of molecular simulations is their high computational cost. To study procesess that take place in long time scales it is necessary to apply special simulation methods. We use and metadynamics and other methods and we endeavour to contribute to their development, for example by application of artificial neural networks.